Chemical reactions
Who can use this feature?
👤 By default, all Admins and Editors (but not Viewers).
🏢 Available on all plans.
You can add 2D structures right in your experiment summary. Simply drag the file into the editor, or type “/chemical reaction” and press Enter.
This will insert a preview image, while still allowing you to access the original file (hover the image, click the three-dot menu and choose Download).
Users can create a chemical reaction block on Colabra, which supports ability to upload, visualize, and edit chemical structures and chemical reactions. Chemicals can also be searched by name and CAS number.

Supported formats
MDL Molfile
RXN
InChI String
ChemAxon Extended SMILES
ChemAxon Extended CML
Chemical structure search
You can filter down any experiment list to those that include a particular chemical structure based on its SMILES string. You can also create a filtered view for quick access to an up-to-date list of experiments using a chemical.
Video overview
Using the Chemical Structure Editor
Using the Tool palette, you can:
Draw and edit a molecule or reaction by clicking on and dragging atoms, bonds, and other elements provided with the buttons on the Atoms toolbar and Tool palette
Delete any component of the drawing (atom or bond) by clicking on it with the Erase tool
Delete the entire molecule or its fragment using the Lasso, rectangle, or fragment selection and the Erase tool
Draw special structures (see the following sections)
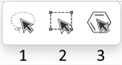
To select one atom or bond, click Lasso (1) or Rectangle Selection tool (2) and click the atom or bond.
To select the entire structure:
Select the Fragment Selection tool (3) and then click the object.
Select the Lasso or Rectangle Selection tool, and then drag the mouse to select the object.
Ctrl-clickwith the Lasso or Rectangle Selection tool.
To select multiple atoms, bonds, structures, or other objects, do one of the following:
Shift-clickwith the Lasso or Rectangle Selection tool selects some (connected or not) atoms/bonds.With the Lasso or Rectangle Selection tool, click and drag the mouse around the atoms, bonds, or structures that you want to select.
Note : Ctrl+Shift-click each structure with the Lasso or Rectangle Selection tool selects several structures.
You can use the buttons of the main toolbar:
Clear Canvas (1) button to start drawing a new molecule; this command clears the drawing area;
Open… (2) and Save As… (3) buttons to import a molecule from a molecular file or save it to a supported molecular file format;
Undo / Redo (4), Cut (5), Copy (6), Paste (7), Zoom In / Out (8), and Scaling (9) buttons to perform the corresponding actions;
Layout button (10) to change the position of the structure to work with it with the most convenience
Clean Up button (11) to improve the structure's appearance by assigning them uniform bond lengths and angles.
Aromatize / Dearomatize buttons (12) to mark aromatic structures (to convert a structure to the Aromatic or Kekule presentation)
Calculate CIP button (13) to determine R/S and E/Z configurations
Recognise Molecule button (16) to recognise a structure in the image file and load it to the canvas (not available in Standalone Mode)
3D Viewer button (17) to open the structure in the three-dimensional Viewer
Help button (19) to view Help Guide
About button (20) to display version and copyright information of the program.
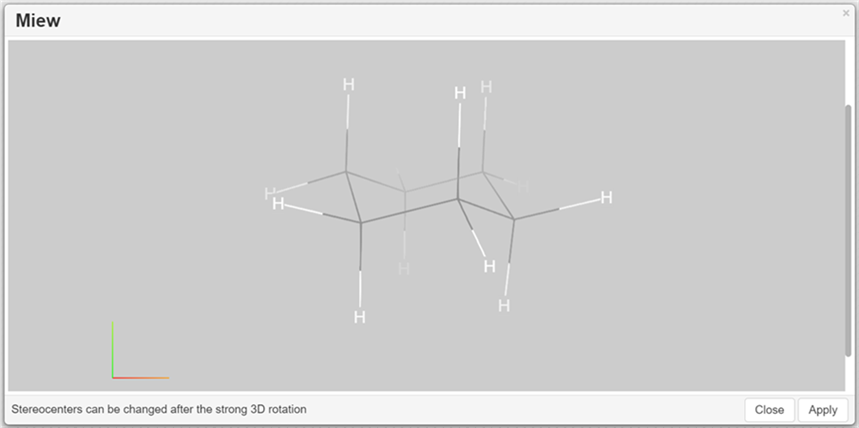
3D Viewer
The structure appears in a modal window after clicking on the 3D Viewer button:
You can perform the following actions:
Rotate the structure holding the left mouse button
Zoom In/Out the structure
Change the appearance of the structure and background colouring.
"Lines" drawing method, "Bright" atom name colouring method and "Light" background colouring are default.
Drawing Atoms
To draw/edit atoms, you can:
Select an atom in the Atoms toolbar and click inside the drawing area
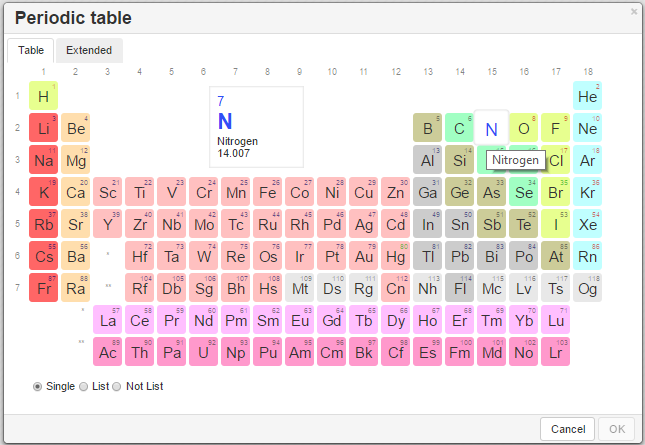
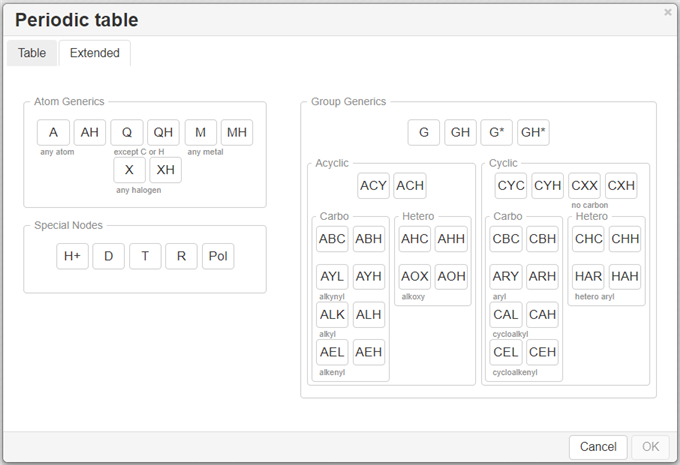
If the desired atom is absent in the toolbar, click on the
button to invoke the Periodic Table and click on the desired atom.
The available options→ Single – selection of a single atom, list – choose an atom from the list of selected options to allow one atom from a list of atoms of your choice at that position, Not list - exclude any atom on your list at that position.
Add an atom to the existing molecule by selecting an atom in the Atoms toolbar, clicking on an atom in the molecule, and dragging the cursor; the atom will be added with a single bond; vacant valences will be filled with the corresponding number of hydrogen atoms
Change an atom by selecting an atom in the Atoms toolbar and clicking on the atom to be changed; in the case a wrong valence appears, the atom will be underlined in red
Drawing Bonds
To draw/edit bonds you can:
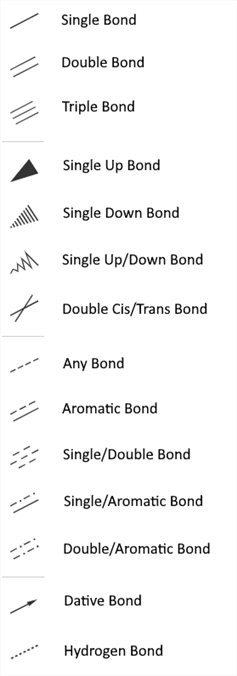
Click an arrow on the Bond tool (Single Bond) in the Tool palette to open the drop-down list with the following bond types:
For the full-screen format, the Bond tool from the Tool palette splits into four: Single Bond, Single Up Bond, Any Bond and Dative Bond, which include the corresponding bond types:
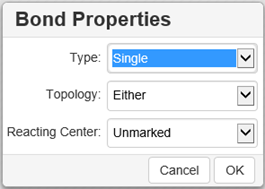
Select a bond type from the drop-down list and click inside the drawing area; a bond of the selected type will be drawn
Adding a bond → click the arrow in the right-hand bottom corner of the Bond tool and select a bond type.
Changing a stereo bond's direction → click on it to automatically change its direction.
Adding a bond to an atom → click on the atom, the bond will be added at a 120º angle.
Adding a bond to an existing molecule → click on an atom in the molecule and drag the cursor. You will be able to choose the degree angle.
Drawing consecutive bonds → use the Chain tool, you can change the atom type
Clicking on the drawn stereo and dative bonds changes their direction
Clicking with the Single Bond tool or Chain tool switches the bond type cyclically: Single-Double-Triple-Single.
Drawing R-Groups
Use the R-Group toolbox
to draw R-groups in Markush structures:
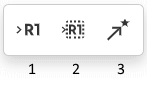
Selecting the R-Group Label Tool (1) and clicking on an atom in the structure invokes the dialogue to select the R-Group label for a current atom position in the structure:
Selecting the R-Group label and clicking OK converts the structure into a Markush structure with the selected R-Group label:
Note: You can choose several R-Group labels simultaneously:
Particular chemical fragments that may be substituted for a given R-Group form a set of R-Group members. R-Group members can be any structural fragment, including functional groups and single atoms or atom lists.
To create a set of R-Group members:
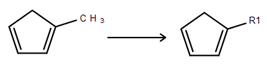
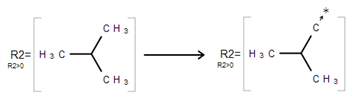
Draw a structure to become an R-Group member.
Select the structure using the R-Group Fragment Tool (2) to invoke the R-Group dialogue; in this dialogue, select the R-Group label to assign the fragment to.
Click on OK to convert the structure into an R-Group member.
An R-Group attachment point is the atom in an R-Group member fragment that attaches the fragment to the initial Markush structure.
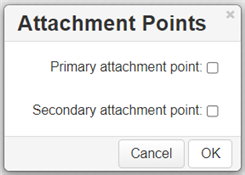
Selecting the Attachment Point Tool (3) and clicking on an atom in the R-Group fragment converts this atom into an attachment point. If the R-Group contains more than one attachment point, you can specify one of them as primary and the other as secondary. You can select between either the primary or secondary attachment point using the dialogue that appears after clicking on the atom:
If there are two attachment points on an R-Group member, there must be two corresponding attachments (bonds) to the R-Group atom with the same R-Group label. Clicking on OK in the above dialogue creates the attachment point.
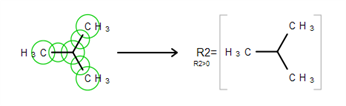
Schematically, the entire process of the R-Group member creation can be presented as:
R-Group Logic
To access the R-Group logic:
Create an R-Group member fragment as described above.
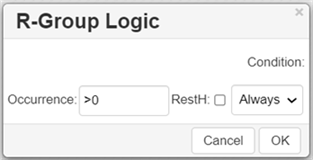
Move the cursor over the entire fragment for the green frame to appear, then click inside the fragment. The following dialogue appears:
Specify Occurrence to define how many of an R-Group occurs. If an R-Group atom appears several times in the initial structure, you will specify Occurrence">n", n being the number of occurrences; if it occurs once, you see "R1 > 0".
Specify H at unoccupied R-Group sites ( RestH ): check or clear the checkbox.
Specify the logical Condition. Use the R-Group condition If R(i) Then to specify whether an R-Group's presence depends on the presence of another R-Group.
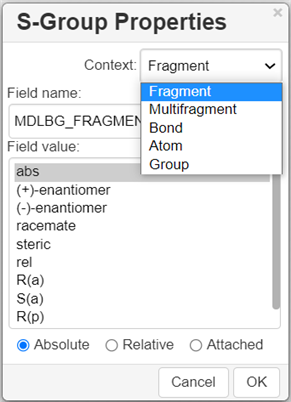
Marking S-Groups
To mark S-Groups, use the S-Group tool
and the following dialogue that appears after selecting a fragment with this tool:
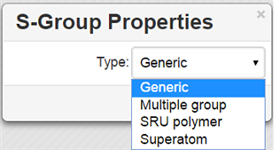
Available S-Group types:
Generic→ Generic is a pair of brackets without any labels.
Multiple group→A' Multiple group' indicates several replications of a fragment or a part of a structure in contracted form.
SRU Polymer→The Structural Repeating Unit (SRU) brackets enclose the structural repeating of a polymer. You have three available patterns: head-to-tail (the default), head-to-head, and either/unknown.
Superatom→ An abbreviated structure (abbreviation) is all or part of a structure (molecule or reaction component) that has been abbreviated to a text label. Structures that you shorten keep their chemical significance, but their underlying structure is hidden. The current version can't display contracted structures but correctly saves/reads them into/from files.
Data S-Groups
The Data S-Groups Tool
is a separate tool for comfortable use with the accustomed set of descriptors (like Attached Data in Marvin Editor).
You can attach data to an atom, a fragment, a multi fragment, a single bond, or a group. The defined set of Names and Values is introduced for each type of selected element:
Select the appropriate S-Group Field Name.
Select the appropriate Field Value.
Labels can be specified as Absolute, Relative or Attached.
Changing Structure Display
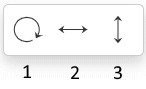
Use the Rotate tool
to change the structure display:
For the full-screen format, the Rotate tool is split into separate buttons:
Rotate Tool (1)
This tool allows rotating objects.
If some objects are selected, the tool rotates the selected objects.
If no objects are selected, or all objects are selected, the tool rotates the whole canvas
The default rotation step is 15 degrees.
Press and hold the Ctrl key for more gradual continuous rotation with a 1-degree rotation step
Select any bond on the structure and click Alt+H to rotate the structure so that the selected bond is placed horizontally. Select any bond on the structure and click Alt+V to rotate the structure so that the selected bond is placed vertically.
Flip Tool (2, 3)
This tool flips the objects horizontally or vertically.
If some objects are selected, the Horizontal Flip tool (or Alt+H) flips the selected objects horizontally
If no objects are selected, or all objects are selected, the Horizontal Flip tool (or Alt+H) flips each structure horizontally
If some objects are selected, the Vertical Flip tool (or Alt+V) flips the selected objects vertically
If no objects are selected, or all objects are selected, the Vertical Flip tool (or Alt+V) flips each structure vertically
Drawing Reactions
To draw/edit reactions you can:
Draw reagents and products as described above;
Use the Reaction Arrow Tool (1) to draw an arrow and pluses in the reaction equation and map the same atoms in reagents and products.
1 – Reaction Arrow Tool
2 – Reaction Arrow Equilibrium Tool
3 – Reaction Plus Tool
4 – Reaction Auto-Mapping Tool
5 – Reaction Mapping Tool
6 – Reaction Unmapping Tool
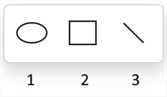
Drawing graphical objects
To draw graphical objects, click the arrow on the Shape Ellipse tool
in the Tools palette to open the drop-down list with the following tools:
Shape Ellipse (1), Shape Rectangle (2), and Shape Line (3).
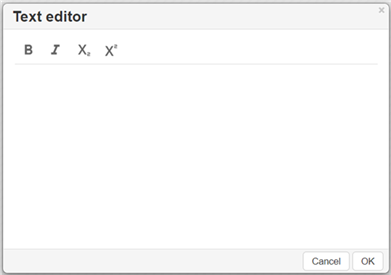
Creating text objects on the canvas
To add text to the canvas, click the Add text tool
in the Tools palette and click the canvas to open the Text editor window:
To enter text, type in the Text editor field.
To edit text, double click the text object on the canvas.
Change the text style to bold and italic, make it subscript and superscript while typing or by selecting text and applying styles.
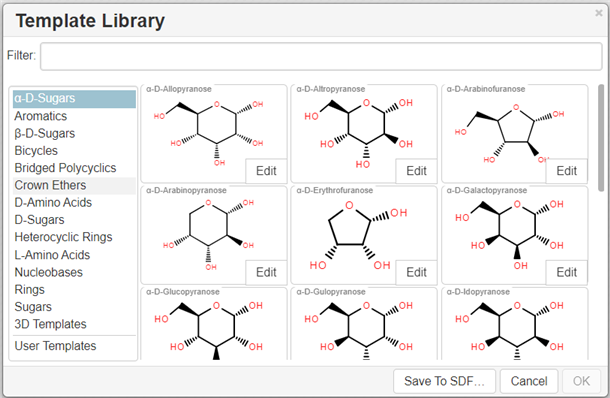
Templates toolbar
You can add templates (rings or other predefined structures) to the structure using the Templates toolbar together with the Custom Templates button located at the bottom:
To add a ring to the molecule, select a ring from the toolbar, click inside the drawing area, or click on an atom or bond.
Rules of using templates:
Dragging the cursor from an atom in the existing structure results in the single bond attachment if the cursor is dragged to more than the bond length; otherwise, the fused structure is drawn. Selecting a template and clicking on a bond in the existing structure created a bond-to-bond fused structure:
The bond in the initial structure is replaced with the bond in the template.
This procedure doesn't change the length of the bond in the initial structure.
Dragging the cursor relative to the initial bond applies the template at the corresponding side of the bond.
Note: The added template will be fused by the program's default attachment atom or bond preset.
Note: The user can define the attachment atom and bond by clicking the Edit button for template structure in the Template Library.
The Custom Templates button
invokes the scrolling list of templates available in the program; both built-in and created by the user:
To create a user template:
Draw a structure.
Click the 'Save as' button.
Click the 'Save to' Templates button.
Enter a name and define the attachment atom and bond.
Last updated